Human Microbiome
Along with other forms decay, it has long been known that micro-organisms are the cause of many harmful and fatal diseases including the bubonic plague, flesh-eating skin disease, tuberculosis, syphilis, cholera, vaginosis and the list continues. It wasn’t until quite recently that scientists began studying the bacteria that commonly make up our microbiomes. Doing so has revealed both the good, the bad, and the opportunistically deviant microbes which cover our bodies and digestive tracts like an over applied cosmetic.
A large ongoing investigation launched back in 2007, branded the “Human Microbiome Project” (HMP), is aimed toward identifying how the human microbiome is associated both with human health and disease, using metagenomic and 16S profiling. Next-generation 16S DNA sequencing techniques have revealed thousands of different bacterial species living in our guts, mouths, skin, airways, bloodstreams, and vaginas. Most of these microbes have had their genomes fully, or partially sequenced using metagenomic profiling.
The purpose of this?
To identify the different species, and genes possessed by the microscopic organisms that very likely consider you their god. Evidence has been shown that the microbes present on your skin and inside our organs at this exact moment, vary greatly from the vaginal or general skin microbiota which dominate your surfaces directly after birth. We can be thankful the microbes that culture our faces, bodies and intestines change steadily from birth until they reach a somewhat stable equilibrium in later life. Evidence is beginning to accumulate on how much plasticity our biomes truly possess. Notably, the mouth and gut microbiomes will achieve the most diversity throughout life.
Studies like HMP have turned up other interesting findings about the microbes which culture various areas of our bodies. It has been found that between individual microbiomes; the species and genes present can vary up 80-90%. This difference is staggering, greatly surpassing the 0.01% difference seen amongst human genomes! The effects of different microbiomes on health and disease have shown correlation between an individual’s microbiome and their metabolism, immune response, acne, antibiotic-associated diarrhea, asthma, allergies, autoimmune diseases, cancer, cavities (dental), depression, anxiety, diabetes, eczema, gastric ulcers, artery hardening, IBS, malnutrition, and obesity. All of this evidence raises the following question, how are these modulatory changes achieved?
Species diversity:
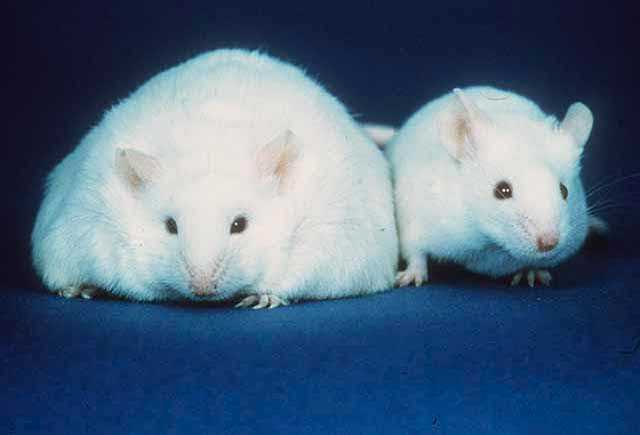
You may have heard that transferring the fecal matter of an obese mouse to a normal mouse, will cause it to also become obese. Well you better believe it, because it’s most definitely the case in several mouse studies dating back over a decade! The root causes of these apparent transformations were followed back to the high abundance of microbes containing diverse metabolic genes which allow them to be exceptionally good at extracting energy from certain foods, as well as microbes involved in signaling the body to store energy as fat within the obese mouse’s stool. It appears that within normal mice the amount of Firmicutes is roughly the same or less than the amount of Bacteroidetes; which are the two major bacterial taxa composing the gut. In the obese mouse, however, the amount of Firmicutes was much higher than the amount of Bacteroidetes. The significance of specie diversity within our microbiomes is becoming very apparent, with possible treatments for IBS, malnutrition and obesity through therapeutic fecal transplantation being looked into.
Microbial protein production
It’s true, the microbes which compose our microbiomes bring their own diverse set of genes to the table, but sometimes they even get a little help from a friend. Virulence factors, resistance factors, and metabolic factors can be exchanged between gut microbiome and foreign microbes. A great example being a recent study done by Hehemann et al., in which Japanese civilian’s gut microbiomes were sequenced. Results showed the presence of DNA coding for a porphyranase enzyme, capable of catabolizing the sulphated polysaccharide porphyrans found in red algae of the genus Porphyra. Intrigued, further studies identified this enzyme thought to be unique to marine bacterium Zobellia galactanivorans, as being transferred to the gut bacterium Bacteroides plebeius of Japanese individuals. This fascinating display of change, shows the adaptability of our gut biomes. By gaining genes from foreign microbes, our microbiomes are able to add to the number of metabolic tools available!
Could this be a form of microevolution? Could scientists one day enable humans to digest common cattle feed; able to breakdown cellulose by the addition of a digestive enzyme through microbial gene transfer? Only time will tell my friends.
Small molecules
For this final section, I hope that you have a burning passion for organic chemistry; because recently, scientists have been after the small molecules synthesized by the human microbiome, identifying their roles in microbe-host and microbe-microbe interactions. Donia et al., describes an array of small molecules that have been isolated from gut microbiota spanning a wide spectrum of chemical classes having antibiotic, neurotransmitter, immunomodulatory and metabomodulatory activities. Several classes have been identified, including:
Ribosomally synthesized posttranslationally modified peptides (RiPPs), are released by bacteria and are toxic to various other species in large enough concentration. Often used to regulate food sources, and “control land from invaders” as our friends to the south would agree with. These modified peptidyl structures are relatively large and span various antibiotic and cytotoxic classes including bacteriocins, lantibiotics, thiopeptides, and microcins.
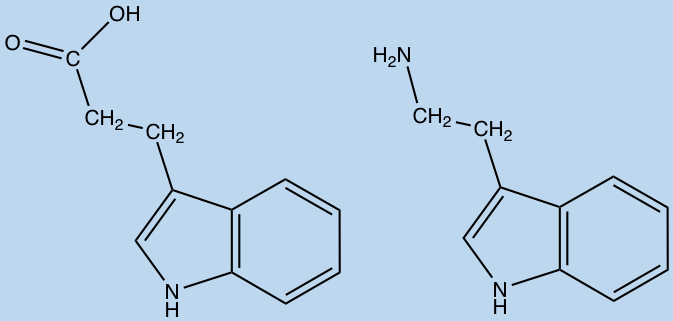
Amino acid metabolites produced by the gut microbiota can be diverse in structure and very few have characterized activity in humans. For instance, the metabolites of tryptophan and phenylalanine; tryptamine and phenethylamine, have been linked to signaling interactions in the enteric nervous system. While others such as indolepropionic acid, p-cresol and phenyllactic acid are currently unknown.
Indolepropionic acid (left) and Tryptamine (right) (ChemDraw)
A “smaller” small molecule class, called the terpenoids, are believed to be created in the human gut by bacteria metabolizing bile acids, producing molecules such as deoxycholic acid which have been proposed to have metabomodulatory effects, however, this class still requires much research.
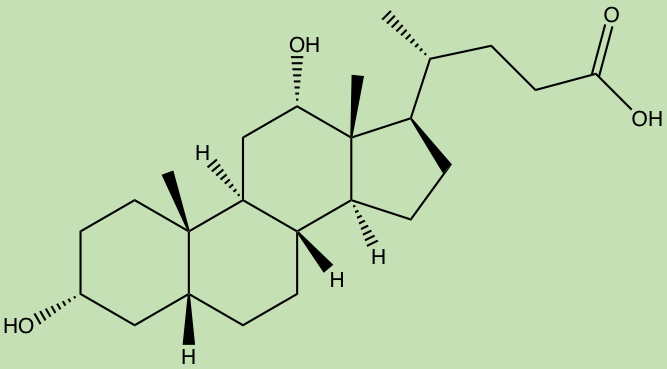
Deoxycholic acid (ChemDraw)
Various glycolipids, glycoproteins and oligosaccharides secreted by microbes have been shown to exhibit immunomodulatory action by activating various NOD and CDId receptors signaling immune responses. These molecules are released for many reasons including beneficial signaling and defense, but may include biofilm formation… Depending upon the strength of the signal, an appropriate immune response, including the recruitment of Natural Killer T-cells to the location of incidence may occur.
The last group of molecules includes, nonribosomal peptides (NRPs) and polyketides (PKs) common to various pathogenic bacteria, however, few associated with the human-microbiome are known. NRPs and PKs such as mycolactone and tilivalline have been identified from pathogenic bacterial species. The molecule mycolactone is a PK secreted by Mycobacterium ulcerans, and is the cause of cell necrosis, immune suppression and eventually ulceration. Klebsiella oxytoca produces the molecule tilivalline, an NRP associated with causing hemorrhagic colitis… Discovering the most abundant and biologically active molecules produced by the microbiota, and connecting them to the genes that encode them, are critical first steps in understanding which molecules have desired effects and which are detrimental. Further studies must be aimed at identifying what receptors these molecules target, and how therapeutic communities of microorganisms can be designed in which the production of molecules indicative of therapeutic action can be specified genetically by altering the microbiome.

Tilivalline (ChemDraw)
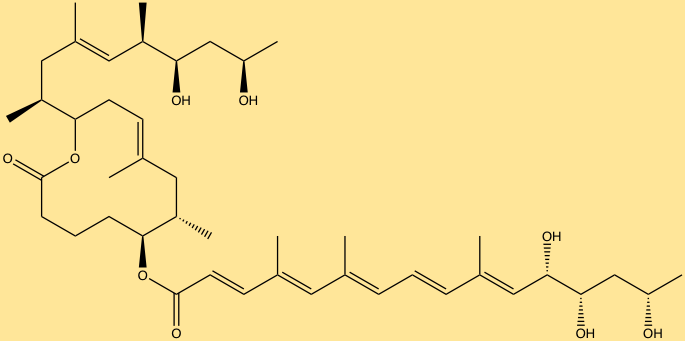
Mycolactone (ChemDraw)
Recent Comments